CellCard Schema Introduction
This web page provides an introduction to the Cell Card Schema and the schema's role in structuring the Cell Card data.
The CellCard Schema is developed using the Linked Data Modeling Language (LinkML). LinkML allows developers to design schemas that are easily shared across many platforms and communities. By using LinkML, we can easily define the minimum information standards, the structure of the cell cards, mappings between cell card fields and ontology terms, and generate multiple types of schemas, such as JSON and SQL schemas, to fit the needs of various information systems. The generated schemas will allow us to validate data that goes into the cell cards and disseminate documentation to the community about the information standards required to produce CellCards. We will also use LinkML to drive the CellCards user interface. This will standardize the workflows, making it easier for other groups to use and ensuring rigor and reproducibility, and facilitate the integration of data from knowledge base created in other resources such as ASCT+B2.
Below is a tutorial on the Cell Card Schema, its usages in our cellcards project, and data validation:
Cell Card Schema and Example
The following figure illustrates how the Cell Card Schema works:
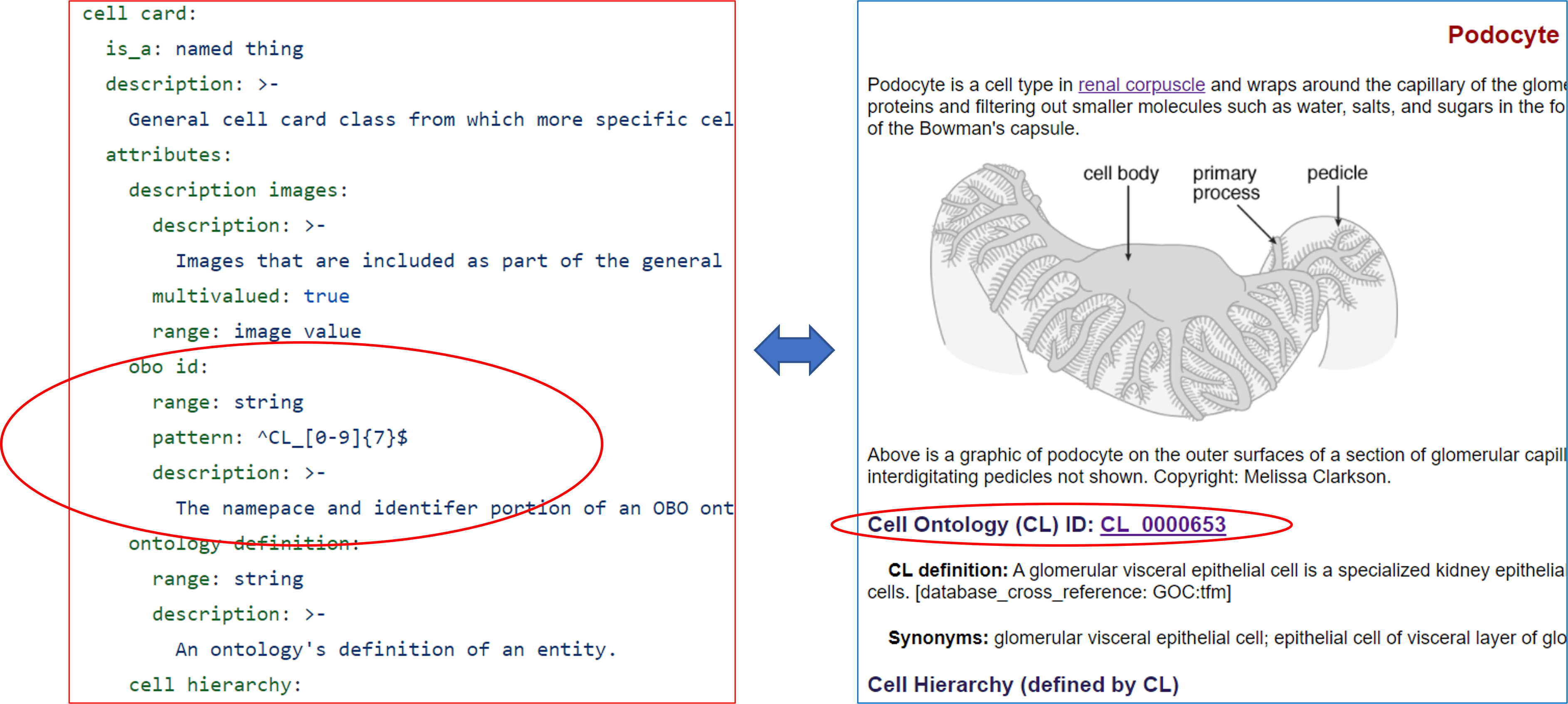
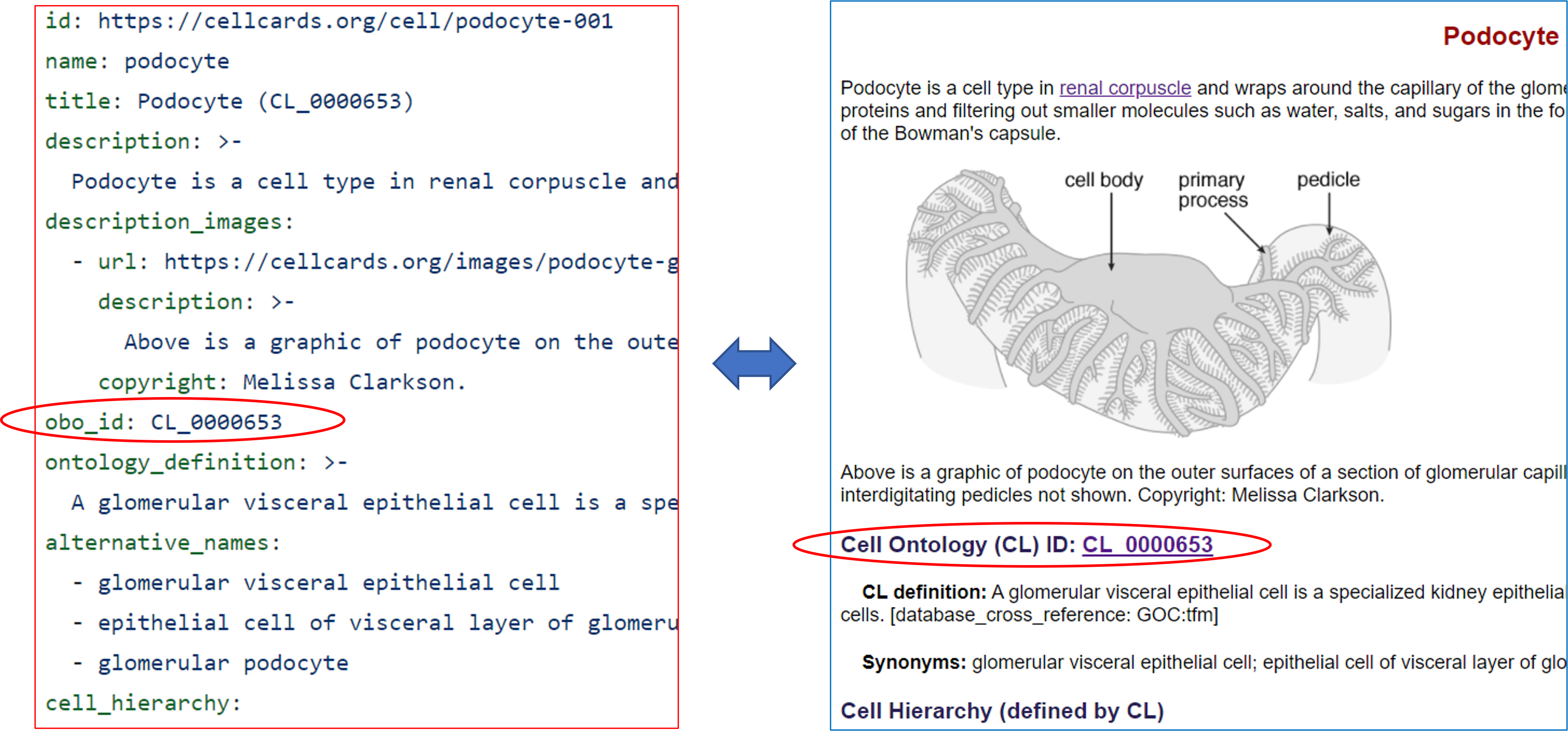
Using the CellCard schema, we can computationally validate data before submitting the data to the information system used to display the data on the CellCard web page. In the example below, we see the podocyte data, represented in YAML (on the left below), and the corresponding podocyte web page (https://cellcards.org/podocyte.php). The CellCard schema (https://github.com/CellCards/CellCard-Schema/blob/main/src/linkml/cellcard.yaml) (as shown on the left above) specifies that an OBO ID must begin with the characters "CL_" followed by 7 digits. An example of data validation is provided in the "Validating CellCard Data" section below. Note, the data does not necessarily have to be structured as YAML. JSON, CSV, and RDF formats are also permitted.
Furthermore, LinkML allows us to automatically create the Cell Card Schema Documentation that provides details on how the schema is defined. The following is an example:
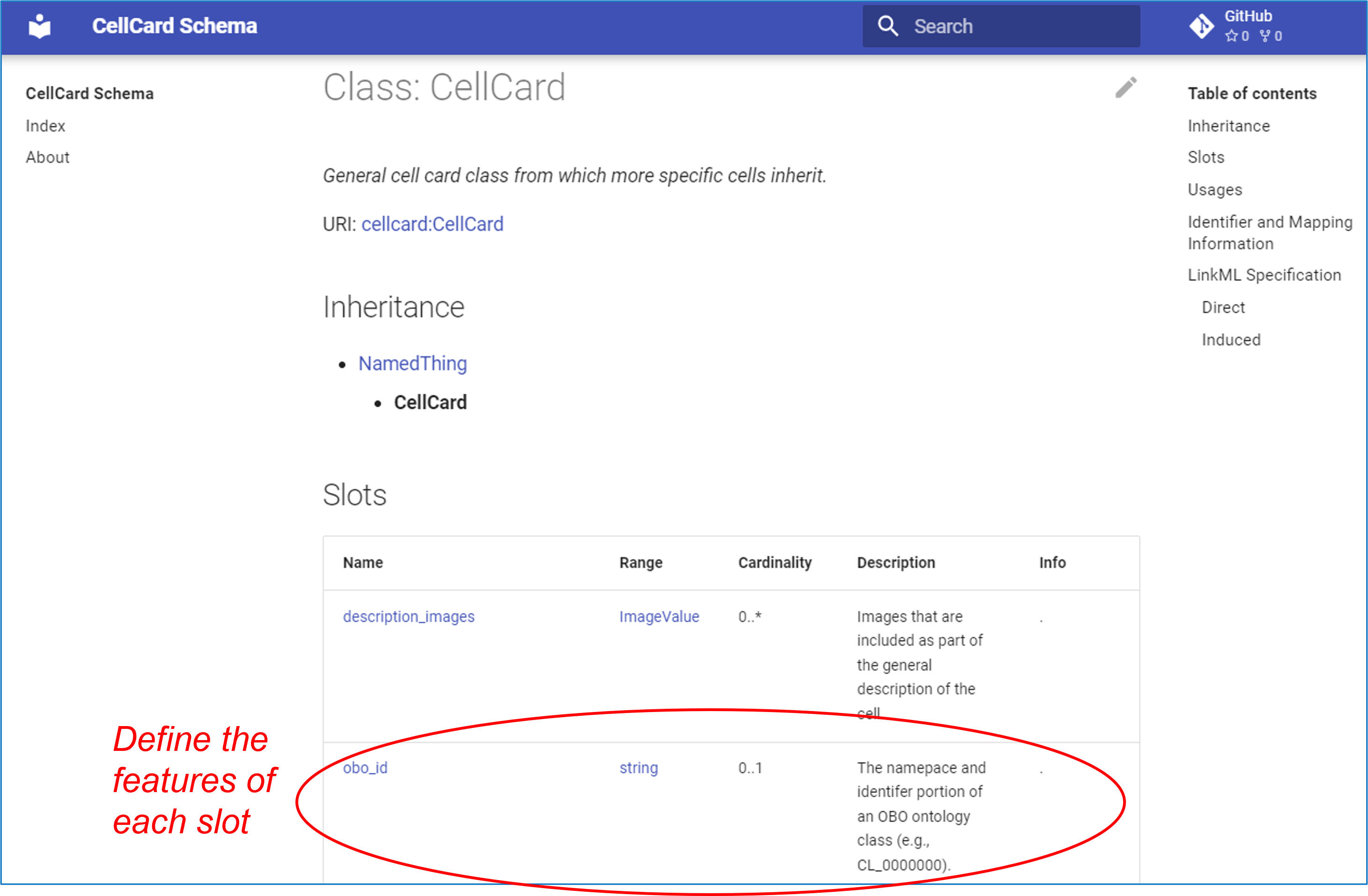
As shown on the above screenshot, the obo_id is defined as a string value. More specific restrictive definition about the obo_id is provided earlier and below.
Validating CellCard Data
We are also able to validate whether specific data are valid based on our cell card schema.
The following Jupyter (Python) notebook script illustrates an example of loading and validating podocyte data.
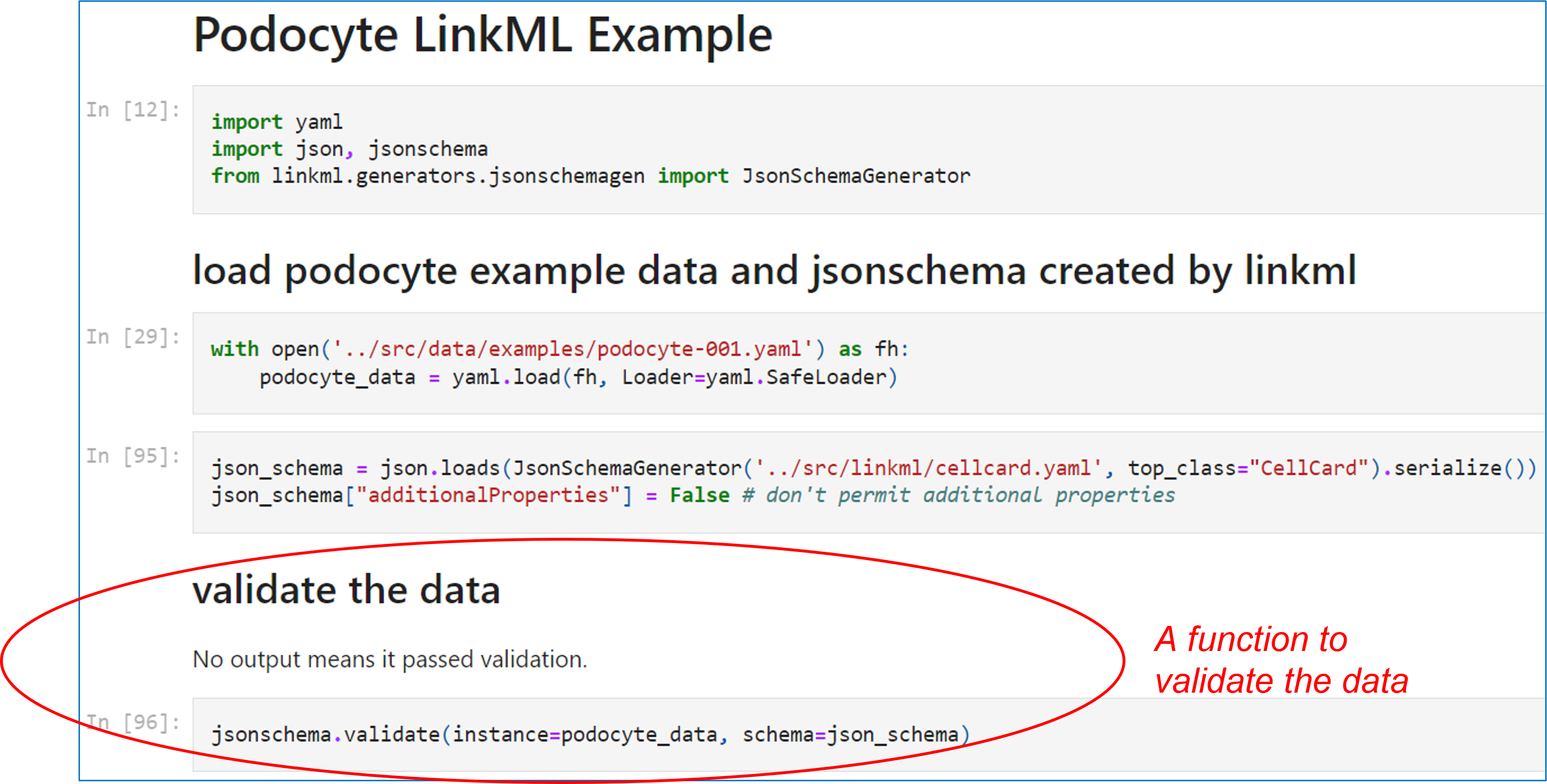
The source of the above notebook is here: https://github.com/CellCards/CellCard-Schema/blob/main/notebooks/podocyte-linkml-example.ipynb.
The following example (from the same notebook source as above) tests some invalid data such as an invalid OBO ID:
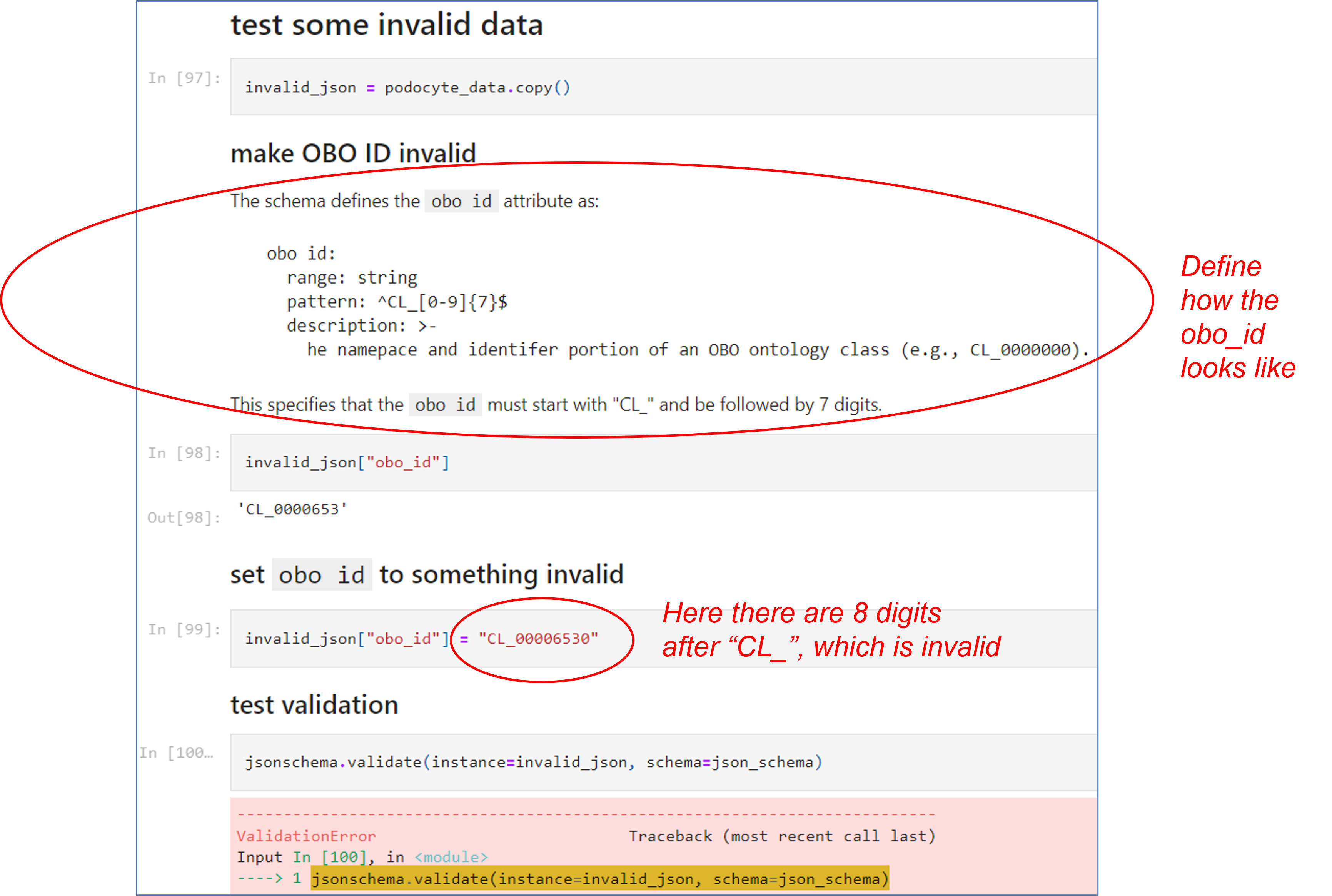
As shown on the above screenshot, the Cell Ontology (CL) obo_id is defined as a string that starts with "CL_" followed with 7 digits. Therefore, the string "CL_0000653" is a valid obo ID; however, "CL_00006530" is not a valid CL obo_id since it has 8 digits rather than 7.
Cell Card Data Generation
There are different ways to generate the cell cards. The podocyte cell card data were manually generated. Later, we can more automatically generate the cell card data using different methods.
One method of cell card data generation is to generate a standard spreadsheet template, whicn can be used by data submitters (or data submitting tools) to generate and submit data for a specific cell card. The populated sheet will be validated by a data validator.
There are different ways to validate the submitted data. For example, the DataHarmonizer (Hsiao Lab) can be used to support the data validation, which can be internally supported by our LinkML-based schema as described above.
A submitted and validated data can be stored in our CellCards server in different ways, faciliting the public query, analysis, and downloading.
Web links introduced above:
- CellCard Schema GitHub (https://github.com/CellCards/CellCard-Schema):
- Schema cellcard.yaml: https://github.com/CellCards/CellCard-Schema/blob/main/src/linkml/cellcard.yaml
- Data example (podocyte): https://github.com/CellCards/CellCard-Schema/blob/main/src/data/examples/podocyte-001.yaml
- Jupyter notebook example (podocyte): https://github.com/CellCards/CellCard-Schema/blob/main/notebooks/podocyte-linkml-example.ipynb
- Schema Documentation:
- CellCard Schema Documentation: https://cellcards.github.io/CellCard-Schema/CellCard/
- Linkml tutorial: https://linkml.io/linkml/intro/tutorial.html
- Podocyte Cell Card: https://cellcards.org/podocyte.php
- DataHarmonizer GitHub: https://github.com/cidgoh/DataHarmonizer.
More information will be provided later. Stay tuned ...